Abstract
INTRODUCTION: Epigenetic dysregulation is a hallmark of Acute Myeloid Leukemia (AML). Although recurrent epigenetic-related mutations have been identified, the understanding of how they affect pathophysiology and leukemogenesis remains incomplete. We used proteomics to assess the epigenetic landscape in AML and identified new prognostic phenotypes and therapeutic targets.
METHODS: Using Reverse Phase Protein Array (RPPA) 405 proteins (327 total; 78 post-translational modified) were measured in 818 samples from newly diagnosed adult AML patients and normalized to CD34+ cells. These included Histone H3 total and H3K4Me 1, 2, 3, K9Me2, K27Me3, K27Ac and K36Me3 Modification Marks (HMM) expression. Categorical clinical variables were compared with Fisher's Exact with simulated p-values (10000 replicates), and continuous ones with Wilcoxon or Kruskal-Wallis tests. Univariate (UV) and Multivariate (MV) models for Overall Survival (OS) and Remission Duration (RD) were built with Cox proportional hazards model (CoxPH). Differentially Expressed Proteins were determined using the Kruskal-Wallis test (p<0.05) and pairwise Dunn's tests with p-values adjusted with the False Discovery Rate method (p<0.05).
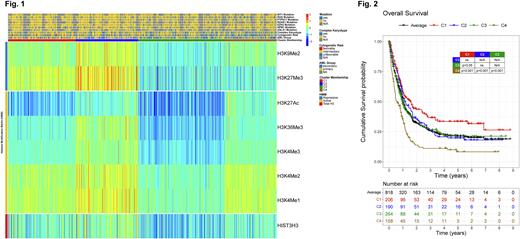
RESULTS: Using unbiased hierarchical clustering, patients were clustered into 4 groups (Fig. 1, C1 to C4) based on the expression of 2 Repressive HMM (H3K27Me3 and H3K9Me2), and 5 Activating HMM (H3K27Ac, H3K36Me3, H3K4Me3, H3K4Me2, H3K4Me1). High repression and low activation HMM were prominent in C1, while C4 had the opposite pattern. Higher expression of all marks and Total H3 defined C2, and C3 had lower expression of all marks, and Total H3. Importantly, when normalized by Total H3, HMM expression of both C2 and C3 are close to average, representing a chromatin state between C1 and C4. Compared to the other groups, patients in C4 were older (66yo vs 62yo; p=0.006), and had more secondary AML (55% vs 43%; p=0.007) and unfavorable cytogenetic risk (55% vs 37%; p<0.001). Mutations weren't exclusively associated with any cluster: ASLX1 and NPM1 mutants were more frequent in C1 and C2 (27% vs 13%; p<0.001; 23% vs 15%; p=0.015), WT1 more in C1 and C3 (9% vs 3.7%, p=0.018), and EZH2, PTPN11, RAS and RUNX1 happened more in C3 and C4 (7.7% vs 1.7%, p<0.001; 13% vs 5.6%, p=0.002; 26% vs 14%, p<0.001; 22% vs 14%, p=0.015).
The effect of HMM expression on OS (Fig. 2) showed that patients with high repression and low activation (C1) did significantly better (Median OS (MS) = 19mo, Median RD (MRD)>102mo), and those with the opposite pattern of low repression and high activation (C4) did significantly worse (MS=7mo, MRD=16mo). Patients with an intermediary chromatin state were close to the average OS (C2: MS=14mo, MRD=20mo; C3: MS=12mo, MRD=24mo). In the UV CoxPH model, C1, C3 and C4 predicted OS (HR=1, 1.3, 1.4, 2.3; p=ref., 0.007, 0.001), as did age, secondary AML, complex karyotype, and CEBPA, FLT3, KIT, JAK2, NPM1, PTPN11, SRSF2 and TP53 mutations. In the MV model C1, C3 and C4 were again predictive for OS (HR=1, 1.5, 2.4, 2.7; p=ref., 0.003, 0.006), together with age, secondary AML, and PTPN11, SRSF2 and TP53 mutations. For RD, in the UV model, all clusters predicted RD (HR=1, 1.9, 1.8, 2.7; p=ref., 0.002, 0.005, 0.001), as did age, secondary AML, complex karyotype, and FLT3, NPM1, RUNX1 and TP53 mutations. In the MV model of RD, all clusters were predictors (HR=1, 1.8, 1.8, 2.7; p=ref., 0.045, 0.038, 0.003), together with age, complex karyotype and RUNX1 mutation. Differentially Expressed Proteins, from the other 397 proteins, for C4 revealed high activation of VEGF, ErbB and FcεRI signaling pathways, unveiling potential new druggable targets.
CONCLUSION: HMM expression found that opposing chromatin states, with a more closed (C1) or open (C4) chromatin, significantly affected prognosis. Moreover, the epigenetic landscape was useful to predict OS and RD: compact chromatin equals high OS and RD (C1), and a loose one means poor OS and RD (C4), whereas an intermediary state (C2/C3) leads to average OS and RD. The expression pattern of the worst OS group showed high activation of specific signaling pathways which could be targeted to improve OS and RD. It would be worthwhile to correlate the findings with ATAC-seq, ChIP-seq, RNA-seq and DNA methylation analysis, in order to determine the loci responsible for the observed phenotypes in each cluster, and also WGS to identify possible new drivers of chromatin aberrations in AML.
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.